Cufflinks Rna Seq Tutorial
Test_chromosome Cufflinks exon 351 400 1000. RNA-Seq Tutorials Tutorial 1 - RNA-Seq experiment design and analysis - Instruction on individual software will be provided in other tutorials Tutorial 2 - Hands-on using TopHat and Cufflinks in Galaxy Tutorial 3 - Advanced RNA-Seq Analysis topics.
Tophat Cufflinks Command Pipeline
Using TophatCufflinksedgeR to analyze RNAseq data Step 1.
Cufflinks rna seq tutorial. CBSU RNASeq pipeline options file. In this lab you will be working with paired-end reads 2x50 nt that derive from human brain polyA RNA and were produced on the Illumina HiSeq 2000 instrument. 3 Pipelines for RNA-seq RNA-seq analysis pipeline Best practices on the differential expression analysis of multi-species RNA-seq - Genome Biology 2021.
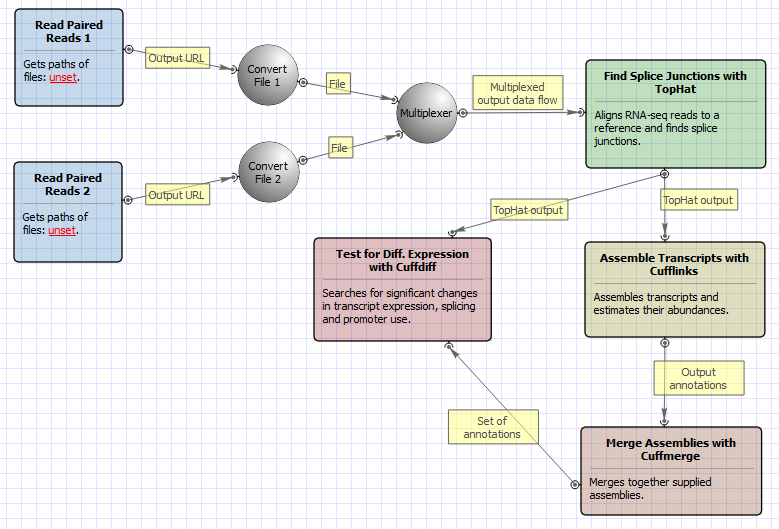
Br h2 Common uses of the Cufflinks package br p Cufflinks includes a number of tools for analyzing RNA-Seq experiments. It accepts aligned RNA-Seq reads and assembles the alignments into a parsimonious set of transcripts. 6 of 14 STEP 2 MERGING THE CUFFLINKS OUTPUT FILES USING CUFFMERGE In Step 2 the output files from the Cufflinks runs transcriptsgtf are merged to create a consensus file that provides the basis for calculating gene and transcript expression for.
Using these tools will allow you to identify new genes and transcripts and then analyse them for differential expression. Bioinformatics Tutorial - Basic. RNA-Seq Tutorials Tutorial 1 RNA-Seq experiment design and analysis Instruction on individual software will be provided in other tutorials Tutorial 2 Advanced RNA-Seq Analysis topics Hands-on tutorials Analyzing human and potato RNA-Seq data using Tophat and Cufflinks in Galaxy.
S_1_sequencetxt and s_5_sequencetxt. A set of lectures in the Deep Sequencing Data Processing and Analysis module will cover the basic steps and popular pipelines to analyze RNA-seq and ChIP-seq data going from the raw data to gene lists to figures. Expression Analysis Of Rna-Seq Experiments With Tophat And Cufflinks.
Much of Galaxy-related features described in this section have been developed by Björn Grüning bgruening and configured. These lectures also cover UNIXLinux commands and some programming elements of R a popular freely available statistical software. Test_chromosome Cufflinks exon 501 550 1000.
On the sample preparation and sequencing protocols that were used. RNA-Seq Tutorials Tutorial 1 RNA-Seq experiment design and analysis Instruction on individual software will be provided in other tutorials Tutorial 2 Hands-on using TopHat and Cufflinks in Galaxy Tutorial 3 Advanced RNA-Seq Analysis topics. 85 years ago by.
Cufflinks assembles transcripts estimates their abundances and tests for differential expression and regulation in RNA-Seq samples. A very comprehensive tutorial of edgeR can be found in. RNA-Seq Tutorial 1 John Garbe Research Informatics Support Systems MSI March 19 2012.
Gene annotation file in gff format Rnaseqconf. RMATS MISO Cufflinks. It also covers several accessory tools and utilities that aid in.
A newer more advanced worfklow was introduce with Cufflinks version 220 and is shown on the right. Reference-Based RNA-Seq Data Analysis Workshop Session 2 Exercise. RNA-seq data comes in many flavors depending eg.
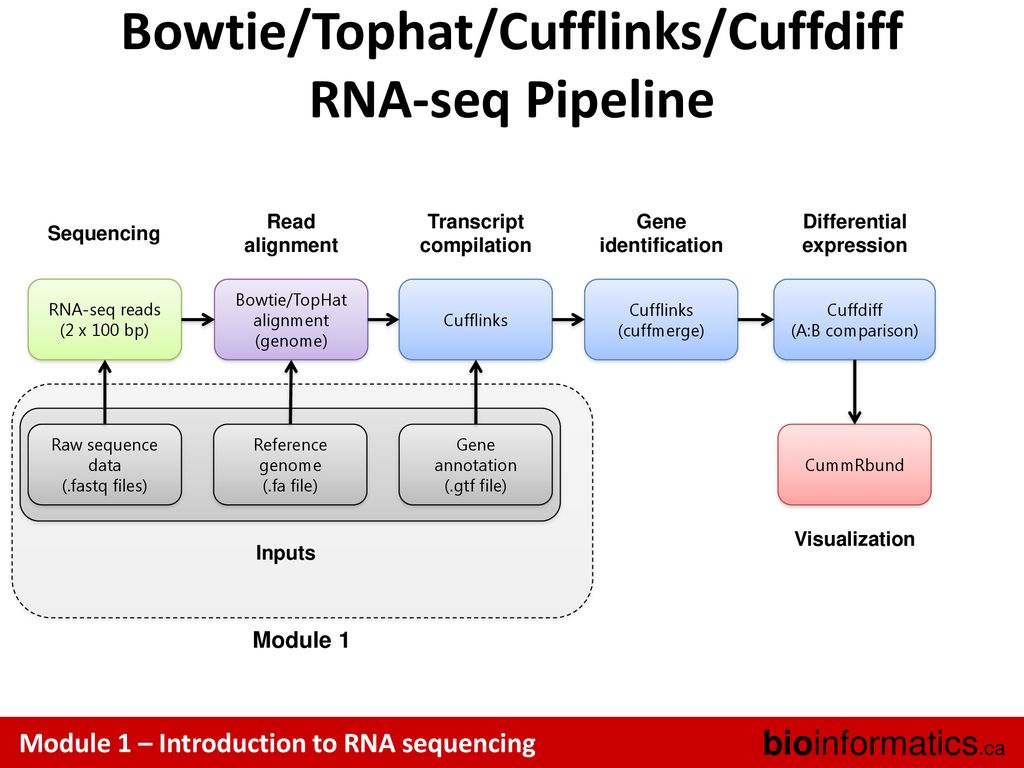
Istvan Albert 85k wrote. Reference-based This tutorial is inspired by an exceptional RNAseq course at the Weill Cornell Medical College compiled by Friederike Dündar Luce Skrabanek and Paul Zumbo and by tutorials produced by Björn Grüning bgruening for Freiburg Galaxy instance. The left side illustrates the classic RNA-Seq workflow which includes read mapping with TopHat assembly with Cufflinks and visualization and exploration of results with CummeRbund.
Cufflinks and Cuffmerge can build a new reference transcriptome by identifying novel transcripts and genes in the RNA-seq dataset - ie. This protocol describes in detail how to use TopHat and Cufflinks to perform RNA-seq analyses. Maize genome sequence in FASTA format ZmB73_5a59_WGSgff.
Illumina data files in FASTQ format maizefa. Cufflinks then estimates the relative abundances of these transcripts based on how many reads support each one. One of CBSU BioHPC Lab workstations has been allocated for your workshop exercise.
Some of these tools can be run on their own while others are pieces of a. Gene Codes Corporation 2017 RNA-Seq p. Istvan Albert 85k.
Canadian Bioinformatics Workshops Ppt Download
Next Generation Sequencing Rna Seq Unix Tools And Igv
The Cufflinks Rna Seq Workflow

Rna Seq Analysis With Tuxedo Tools Unipro Ugene User Manual V 34 Wiki